Strudel
Strudel is our graphical tool for visualizing genetic and physical maps of genomes for comparative purposes. The application aims to let the user examine their data at a variety of different levels of resolution, from entire maps to individual markers, and explore syntenic relationships between genomes. All browsing and interaction with Strudel happens in real-time – there is no need to wait while the maps are generated. It is built using Java 1.6 and ships with its own JRE, so there is no need for users to install or update Java.
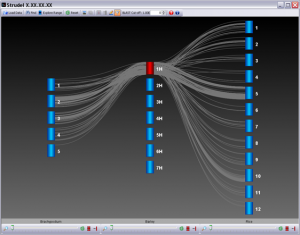
This application is free to download and use but no warranty is given as to its correct functioning.
Features:
- Visualize multiple genomes and thousands of homologies interactively and in real time
- Continuous zoom ranging from entire genome views to singles features
- Explore intervals such as QTLs and list annotation for features
- Search for features by name
- Filter homologies on display by BLAST e-value
- Single genome mode also supported
- Ships with example dataset to facilitate familiarization with the software
Application use cases
We are aiming to release a number of successful use cases of Strudel here as they develop. The first of these relates to the investigation of the Intermedium-C gene in barley which lead to a publication in Nature Genetics (Ramsay et al 2011 Nature Genetics 43(2) 169-172 ) and involved the use of Strudel for the investigation of homologous regions in rice and in Brachypodium distachyon. This is available here as a pdf formatted presentation.
